Nanofabulous Seminar: DNA origami for biotechnological applications
DNA origami has enabled the construction of DNA nanostructures with unprecedented structural control over size, shape and surface functionality. Using one long strand of ssDNA, and hundreds of short DNA oligos, user-defined complex structures can be formed via sequence-dependent self-assembly in a one-pot annealing reaction. Shapes such as cubes, bricks, cylinders and triangles can be formed, all the way to programmed higher ordered assemblies, molecular rotors, transport pores, and logic gates. Importantly, DNA origami objects are proving to be useful tools in an array of applications such as sensing, nanoplasmonics, nanophotonics and drug delivery.[1]
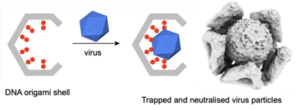
In this seminar, recent work in the development of DNA origami for biotechnological applications will be discussed. First, the use of functionalised DNA origami shells for virus trapping and neutralisation will be presented. Where, upon binding to the interior of the DNA shells, the viruses are blocked from undergoing interactions with host cells.[2, 3] Secondly, recent results demonstrating the genetic encoding of DNA origami for mammalian gene expression will be discussed.[4, 5]
References
[1] P. Wang, et al., Chem, 2017, 2 (3), 359–382.
[2] C. Sigl, et al., Nat. Mater., 2021, 20 (9), 1281–1289.
[3] A. Monferrer*, J. A. Kretzmann*, et al., ACS Nano, 2022, 16 (12), 20002 – 2009.
[4] J. A. Kretzmann, et al., Nat. Commun., 2023, 14 (1), 1017.
[5] A. Liedl, et al., J. Am. Chem. Soc., 2023, 145 (9), 4946 – 4950.
Dr Jessica A. Kretzmann
Forrest Fellow, School of Molecular Sciences
The University of Western Australia, Crawley, WA, Australia
11:00am, 21/09/2023
Melbourne Centre for Nanofabrication Boardroom
151 Wellington Road, Clayton, 3168
Zoom link: click here
Meeting ID: 821 4587 5296 and passcode: 551496


